Welcome to the Atlas of Biosynthetic Gene Clusters in the Human Microbiome!
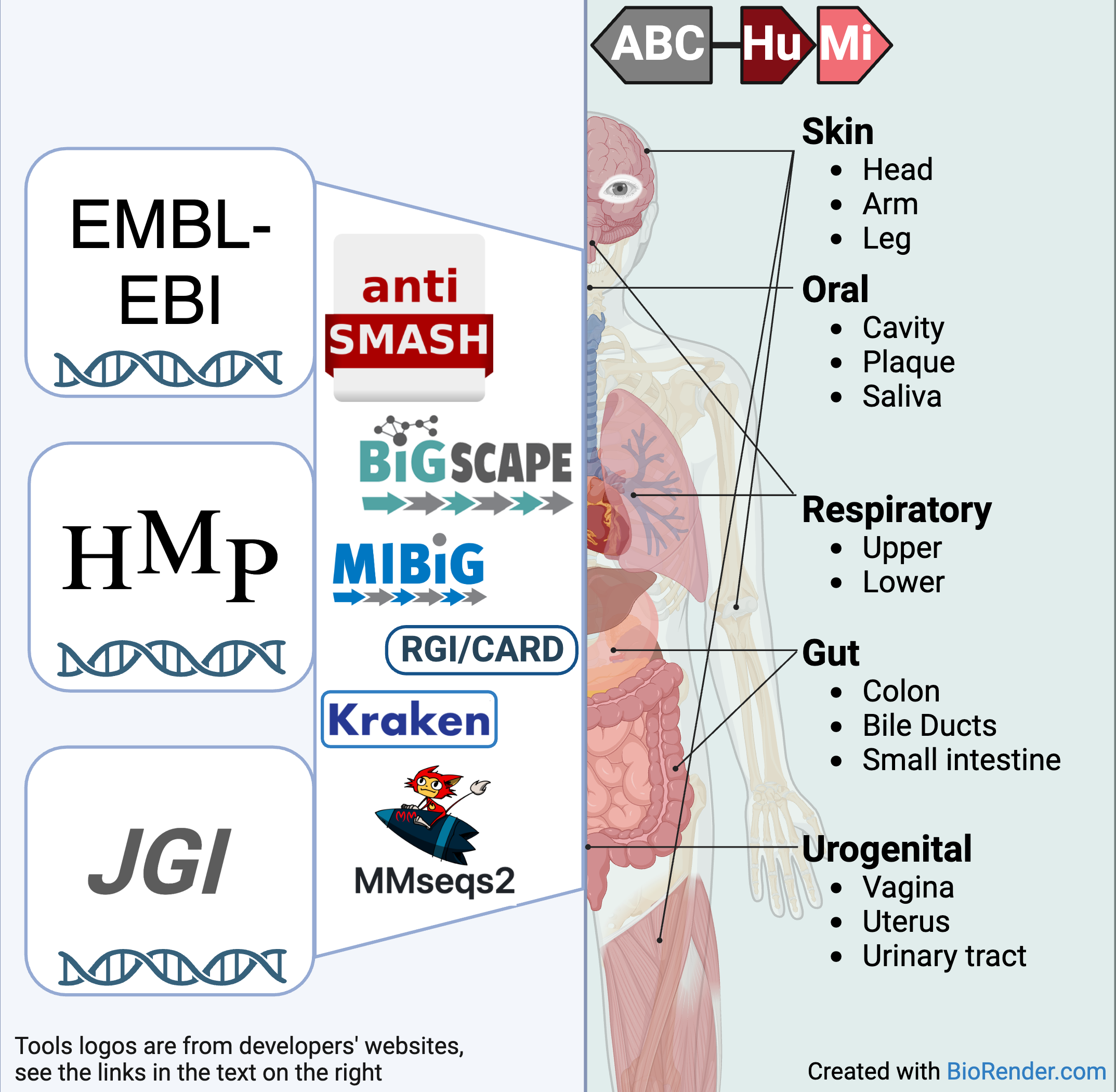
The ABC-HuMi database is an online collection of biosynthetic gene clusters (BGCs) that putatively encode specialized metabolites in the human microbiome. The BGCs were computationally predicted from metagenomic samples and representative prokaryotic genomes collected from 14 body sites of five major groups: Gut, Oral, Skin, Respiratory and Urogenital systems.
The (meta)genome sequences were downloaded from major human microbiome sequence databases, including the JGI, EMBL-EBI, and HMP. We used antiSMASH (v.7.0.0) for BGC mining and BiG-SCAPE (v.1.1.5) for clustering them into gene cluster families (GCFs) and clans (GCC). MIBiG (v.3.1), the largest database of experimentally validated BGCs, was used to determine the novelty of the predicted gene clusters. Our data processing pipeline also relies on Kraken2 (taxonomic classification for the HMP data), MMseqs2 (deduplication), and RGI/CARD (prediction of antibiotic resistance genes).
You can view and filter database entries on the Browse page, which also offers numerous interactive visualizations of the database statistics. Head to the Query page to seamlessly search for custom genes or entire BGCs. Navigate to Cytoscape, to visually comprehend BGC similarity networks. Download the entire database or its parts categorized by body site in the GenBank format or as the preprocessed BiG-SCAPE input. Consult with the Help page if you have any problems with the database or simply Contact us. We would also love to hear your feedback and feature suggestions.
The database is maintained by the Helmholtz Institute for Pharmaceutical Research (HIPS) and the Chair for Clinical Bioinformatics, Saarland University.
To cite the ABC-HuMi database, please reference
ABC-HuMi: the Atlas of Biosynthetic Gene Clusters in the Human Microbiome
Hirsch, P., Tagirdzhanov, A., et al. Nucleic Acids Res. (2023), DOI: 10.1093/nar/gkad1086